| A | B |
|---|
| * The enzymes that catalyze the elongation of new DNA at a replication fork by the addition of nucleotides to the existing chain are called _____. | DNA polymerases (the orange molecules in the picture below) pp267 - 269,  |
| ** The enzyme that joins RNA nucleotides to make the primer is called ____. | primase (letter B in the picture below) pp267 - 269,  |
| ** The enzymes that untwists and unzips the double helix of DNA at the replication forks are called ____. | helicases (letter A in the picture below) p314,  |
| ** A protein that functions in DNA replication, helping to relieve strain in the double helix ahead of the replication fork is called ____. | topoisomerase p267 |
| ** An enzyme that hydrolyzes DNA and RNA into their component nucleotides, and is used to cut out damaged DNA during nucleotide excision repair, is called _______. | a nuclease p270 |
| * _____ is a linking enzyme essential for DNA replication. It catalyzes the covalent bonding of the 3' end of a new DNA fragment to the 5' end of a growing chain. Specifically, it bonds the sugar from one nucleotide to the phosphate of another nucleotide. | DNA ligase p269,  |
| * The change in genotype and phenotype due to the assimilation of external DNA by a cell is called ____. | transformation (This use of the word transformation should not be confused with the conversion of a normal animal cell to a cancerous one, discussed on page 199. The type of transformation discussed here is also the most common way to get a foreign piece of DNA, usually a plasmid, into a host organism for genetic engineering. You basically mix in the plasmids with that have a foreign piece of DNA in them, like the plasmids with the jellyfish p-glo gene we will be using to transform bacteria into glowing bacteria as they take in the recombinant plasmid through their cell wall and membrane) p258 |
| ** A short segment of DNA synthesized on a template strand during DNA replication which is joined to other short segments to form the lagging strand is called a(n) _____. | Okazaki fragment p268,  |
| ** The cellular process that uses special enzymes to fix incorrectly paired nucleotides that were not corrected by DNA polymerase is called ____. | mismatch repair p270 |
| ** During DNA replication, _________, are molecules that line up along the unpaired DNA strands, holding them apart while the DNA strands serve as templates for the synthesis of complementary strands of DNA. | single-strand binding proteins (letter D in the picture below) p267,  |
| ** The ______ is the site where the replication of a DNA molecule begins. | origin of replication (In a circular bacterial chromosome, there is only one origin of replication. In the much larger eukaryotic chromosome, there are hundreds of them) p265 |
| * The process of copying a cell's DNA is called _______. | DNA replication p263,  |
| ** The shape of a DNA molecule is referred to as a(n) _____. | double helix p261,  |
| ** The enzyme that catalyzes the lengthening of telomeres in germ cells (and many cancer cells) is called _____. | telomerase (This enzyme keeps the telomeres full length in germ cells so that the full length chromosomes can be passed on to offspring. Somatic cells get shorter and shorter at each cell division and eventually, the shortening of the DNA effects not only the telomeres, which act as buffer, but actual genes, thereby limiting the number of cell divisions that somatic cells can undergo during their lifetime) p271 |
| ** The new strand of DNA that can be made continuously in the 5' to 3' direction is called the _____ strand. | leading p268,  |
| ** The process of removing and then correctly replacing a damaged segment of DNA using the undamaged strand as a guide is called _____. | nucleotide excision repair p270, 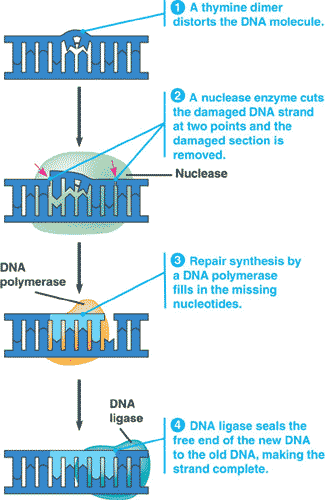 |
| * A virus that infects bacteria is called a(n) _____. | bacteriophage p258, 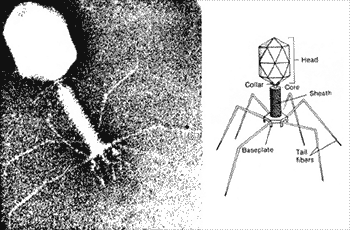 |
| ** The protective structure at each end of a eukaryotic chromosome that consists of repetitive extra DNA. | telomere p271 |
| ** The Y-shaped region on a replicating DNA molecule where new strands are growing is called the ______. | replication fork p266,  |
| ** The discontinuously synthesized DNA strand that elongates in a direction away from the replication fork is called the ______. | lagging strand p268,  |
* The picture below shows a(n) _____.,  | bacteriophage (this virus infects bacteria only) p258,  |
* What type of bonds (represented by the pink dotted lines) hold the bases of complementary DNA together?,  | hydrogen bonds p262,  |
** Which nucleotides below are purines and which are pyrimidines?,  | Thymine and Cytosine are the pyrimidines and Adenine and Guanine are the purines (You might be able to remember that pyrimidines are the ones with a single ring structure because it would be easier to measure the "perimeter" of a single-ring nitrogenous base. Get it? Perimeter, pyrimidine?) p263,  |
| ** According to _____, there is the same percentage of adenine and thymine in the genome of a given species and the same percentage of guanine and cytosine. | Chargaff's rules p260 |
** Which process is shown in the picture below?,  | nucleotide excision repair p270,  |
** The letter A is pointing to the ______., 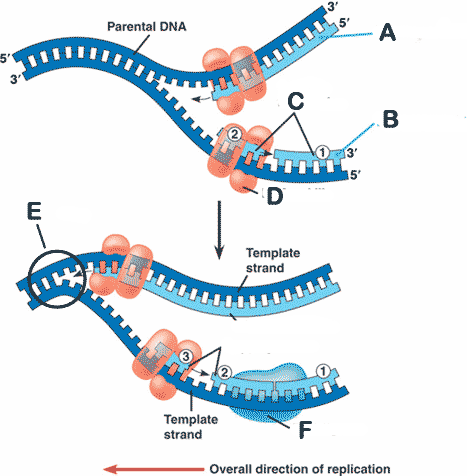 | leading strand pp268 & 269,  |
** The letter B is pointing to the ______.,  | lagging strand pp268 & 269,  |
** The letter C is pointing to the ______.,  | Okazaki fragments pp268 & 269,  |
* The letter D is pointing to ______.,  | DNA polymerase pp268 & 269,  |
* The letter E is pointing to the ______.,  | replication fork pp268 & 269,  |
** The letter F is pointing to ______.,  | DNA ligase (notice that it is catalyzing the bonding of the sugar end of one Okazaki fragment to the phosphate end of the other Okazaki fragment) pp268 & 269,  |
** What is the last name of the scientist(s) who performed the experiment shown below?, 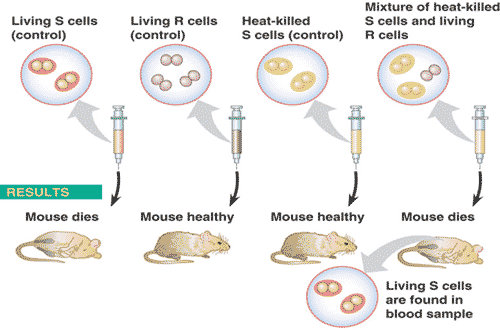 | Griffith (be able to explain this experiment in detail) p258,  |
** What were the last name(s) of the scientist(s) who performed the experiment shown below?,  | Hershey and Chase (be able to explain this experiment in detail) p259,  |
** The letter A is pointing to ______.,  | helicase (this enzyme unwinds and unzips the DNA molecule) pp266 & 267,  |
** The letter B is pointing to ______.,  | primase (This enzyme lays down the necessary primer nucleotides to get the process of DNA replication going. Only one primer is needed on the leading strand while the lagging strand needs a primer for each Okazaki fragment) pp266 & 267,  |
** The letter C is pointing to ______.,  | a primer (This is a short segment of either DNA or RNA that is needed to initiate DNA synthesis because DNA polymerase can only add nucleotides to existing new strands. In humans, the primer is made of RNA nucleotides which will need to be replaced by DNA nucleotides to complete the strand) pp266 & 267,  |
** The letter D is pointing to ______.,  | Single-strand binding proteins (These are proteins that line up along the unpaired DNA strands, holding them apart while the DNA strands serve as templates for the synthesis of complementary strands of DNA.) pp266 & 267,  |
| * The nitrogenous base adenine always bonds to ______ in a DNA molecule. | thymine p263 |
| * The nitrogenous base cytosine always bonds to ______ in a DNA molecule. | guanine p263 |
| * Alfred Hershey and Martha Chase used radioactive ______ to label the proteins in the T2 bacteriophage. | sulfur (They used sulfur because it is found in some amino acids that make up protein, but is never found in the nucleotides that make up DNA) p259,  |
| * Alfred Hershey and Martha Chase used radioactive ______ to label the DNA in the T2 bacteriophage. | phosphorus (They used phosphorus because it is found in the nucleotides that make up DNA, but is never found in the amino acids that make up proteins) p259,  |
| ** Which carbon on the pentose sugar of DNA binds to the nitrogenous base? | 1' p260,  |
| * Which carbon on the pentose sugar of DNA binds to the phosphate group that completes the nucleotide? | 5' p260,  |
| * Which carbon on the pentose sugar of DNA binds to the phosphate group of the next nucleotide in the strand? | 3' (remember that strands are built in the 5' --> 3' direction) p260,  |
** What is this a picture of and what is the name of the technique used to produce it?,  | DNA, X-ray crystallography p261,  |
| ** What were the name(s) of the scientists who first published a paper describing the correct structure of DNA in 1953? | Watson and Crick (They, along with Maurice Wilkins, were awarded the Nobel Prize for their work on determining the structure of DNA. The prize was awarded in 1962. Rosalind Franklin had passed away in 1958 and was therefore ineligible for the prize. Many people believe that Rosalind Franklin did not get the credit she deserved while Watson and Crick got too much.) p257 and pp.260-261 |
| ** What was the name of the scientist(s) who produced images of DNA using X-ray crystallography that helped Watson and Crick decipher the structure of DNA? | Rosalind Franklin (Below is an image of DNA that she produced. Watson and Crick, along with Franklin's partner, Maurice Wilkins, were awarded the Nobel Prize for their work on determining the structure of DNA. The prize was awarded in 1962. Rosalind Franklin had passed away in 1958 and was therefore ineligible for the prize. Many people believe that Rosalind Franklin did not get the credit she deserved while Watson and Crick got too much.) p261,  |
* Which model of DNA replication is correct and what is the model called?, 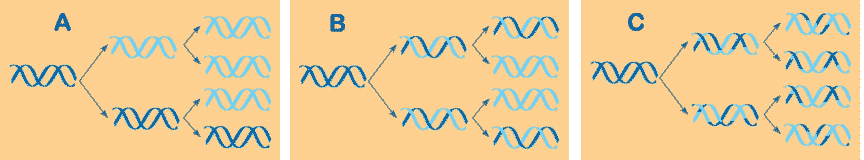 | B) semiconservative model (was fhypothesized by Watson and Crick and experimentally confirmed by Meselson and Stahl using radioactive nitrogen markers to trace the path of the parent DNA strands) pp264 and 265,  |
| ** Which two nucleotides have nitrogenous bases that are classified as pyrimidines? | thymine and cytosine p263, 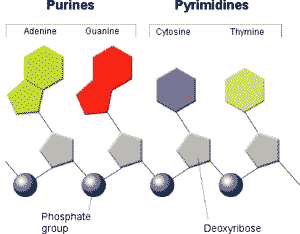 |
| ** Which two nucleotides have nitrogenous bases that are classified as purines? | adenine and guanine p263,  |
| ** Which two complementary pairs of nitrogenous bases bond with 2 hydrogen bonds? | thymine and adenine (You might be able to better remember this by the phrase, "A T for two, GC three") p263, 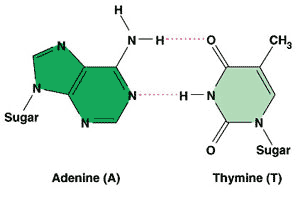 |
| ** Which two complementary pairs of nitrogenous bases bond with 3 hydrogen bonds? | guanine and cytosine (You might be able to better remember this by the phrase, "A T for two, GC three") p263,  |
| * A permanent change in a DNA sequence is called a(n) ______. | mutation p270 |
| * DNA wraps around ________ to form chromatin. | histone proteins p272, 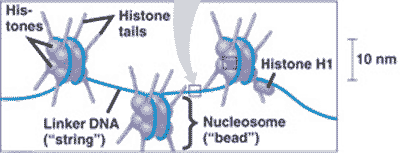 |
* What is "A" pointing to in the picture below?, 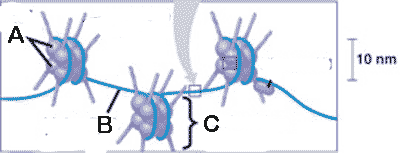 | histone proteins p272,  |
** What is "C" pointing to in the picture below?,  | a nucleosome p272,  |
| ** The more open, unraveled form of eukaryotic chromatin that is available for transcription. | euchromatin p273 |
| ** Nontranscribed eukaryotic chromatin that is so highly compacted that it is visible with a light microscope during interphase. | heterochromatin p273 |
| ** DNA is double stranded, but the two strands run in opposite directions from each other. Because of this, the two strands are said to run _____ to each other.o | antiparallel p263 |
| ** The nitrogenous bases of purines have a ____ - ring structure. | double p263 |
| ** The nitrogenous bases of pyrimidines have a ____ - ring structure. | single p263 |
| ** DNA polmerase can add nucleotides only to the ____ end of a growing DNA strand. | 3' (The apostrophe means "prime" so 3' is translated to "3 prime") |
| * DNA in which nucleotide sequences are combined from two different sources - often different species - is called _______. | recombinant DNA p275, 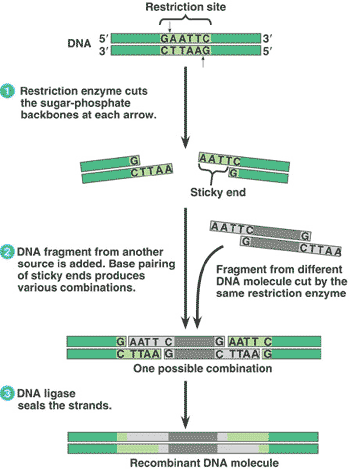 |
| * The direct manipulation of genes for practical purposes is called _____. | genetic engineering p274,  |
| ** Making multiple copies of DNA fragments that code for a specific polypeptide is called _______. | gene cloning p275,  |
| * A common approach to cloning a gene involves splicing the gene of interest into a(n) ____ _____ , then reinserting it back into the ________ which will then reproduce, making many copies of the gene. | bacterial plasmid, bacterium pp274 and 275,  |
| * Enzymes that cut DNA molecules in a limited number of specific locations are called _______. | restriction enzymes p275,  |
| ** The sequence of nucleotides that can be recognized and cut by a particular restriction enzyme is called a(n) ________. | restriction site p275,  |
| ** Pieces of DNA that have been cut by a restriction enzyme are called _______. | restriction fragments p276,  |
| * Restriction fragments (Segments of DNA that were produced using restriction enzymes) can be permanently sealed by an enzyme called _______ which catalyzes the formation of covalent bonds between the sugar at the end of one fragment and the phosphate group at the end of the other fragment. | DNA ligase p276,  |
| ** An agent used to transfer DNA in genetic engineering. Plasmids that move recombinant DNA and viral DNA that can be transfered along with recombinant DNA by infection are both examples. | cloning vector p275,  |
| * A method of quickly producing a lot of DNA copies in vitro from a small amount of original DNA is called ______. | PCR (stands for polymerase chain reactions) p277, 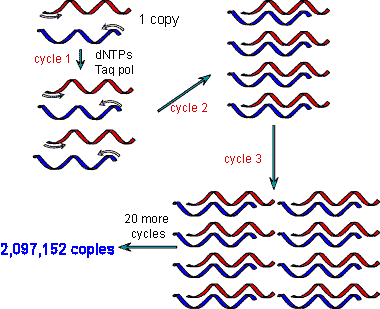 |
| ** Base pairing between a gene and a complementary sequence on another nucleic acid molecule that usually carries some type of marker is called _____. | nucleic acid hybridization (This technique is used to identify genes of interest in a genomic library. A nucleic acid probe is used. This is a short string of nucleotides known to be complementary to a portion of the gene of interest. It has a radioactive isotope, fluorescent marker, or some other type of tag so it can be easily identified and isolated) p274,  |
| * An organism that has acquired one or more genes by artificial means can be called a(n) ______ or a(n) _______. | genetically modified (GMO) organism or a transgenic organism. p.275 |
| * A single-strand end of a double-stranded DNA restriction fragment is called the ______. | sticky end p276,  |
| * A lineage of genetically identical individuals or cells is called a(n) _____. | clone G-6 |
* Which laboratory technique is shown below?, 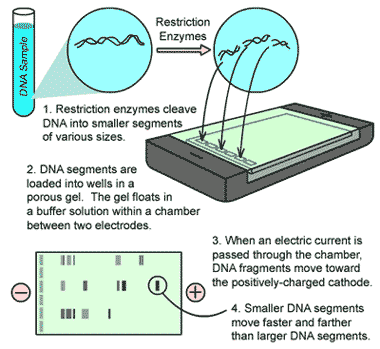 | gel electrophoresis p276,  |
* What is "A" in the picture below?, 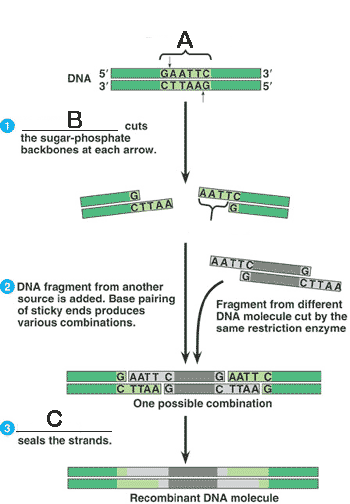 | restriction site p275,  |
* What is "B" in the picture below?,  | restriction enzyme p275,  |
* What is "C" in the picture below?,  | DNA ligase p275,  |
| * Small circular DNA molecules that replicate separately from the main bacterial chromosome are called _____. | plasmids p274,  |
| * _____ is a technique for editing genes in living cells, involving a bacterial protein associated with with a guide RNA complementary to the gene sequence of interest. | CRISPR-Cas9 System p279 |
| ** _____ is a process that biases inheritance such that a particular allele is more likely to be inherited than are other alleles, causing a favored allele to spread through a population. | Gene drive (Scientists are attempting to address the global problem of insect-borne diseases by altering genes in insects so that they can't transmit the disease. They would use gene drive by engineering the new allele so that it is much more highly favored for inheritance than the wild-type allele, allowing it to spread quickly through the population) p280 |